Collections of Neurons#
Last time, we saw how a single neuron was able to “encode” information in its spike train and how we could compute a “decoder”. We use the same LIF and Synapse models as before (but adding an extra reset function for the LIF):
import math
def signal(t):
return 2 * math.sin(t)
class FirstOrderLIF: # First Order Leaky Integrate and Fire
def __init__(self, tau_rc=0.2, tau_ref=0.002, v_init=0, v_th=1):
self.tau_rc = tau_rc # Potential decay time constant
self.v = v_init # Potential value
self.v_th = v_th # Firing threshold
self.tau_ref = tau_ref # Refractory period time constant
self.output = 0 # Current output value
self.refractory_time = 0 # Current refractory period time (how long until the neuron can fire again)
def step(self, I, t_step): # Advance one time step (input I and time step size t_step)
self.refractory_time -= t_step # Subtract the amount of time that passed from our refractory time
if self.refractory_time < 0: # If the neuron is not in its refractory period
self.v = self.v * (1 - t_step / self.tau_rc) + I * t_step / self.tau_rc # Integrate the input current
if self.v > self.v_th: # If the potential is above the threshold
self.refractory_time = self.tau_ref # Enter the refractory period
self.output = 1 / t_step # Emit a spike
self.v = 0 # Reset the potential
else: # If the potential is below the threshold
self.output = 0 # Do not fire
return self.output
def reset(self): # Reset the neuron to its initial state
self.v = self.output = self.refractory_time = 0
class FirstOrderSynapse:
def __init__(self, tau_s=0.01):
self.tau_s = tau_s # Synaptic time constant
self.output = 0 # Current potential
def step(self, I, t_step):
self.output = self.output * (1 - t_step / self.tau_s) + I * t_step / self.tau_s # Decay potential
return self.output
def reset(self): # Reset the synapse to its initial state
self.output = 0
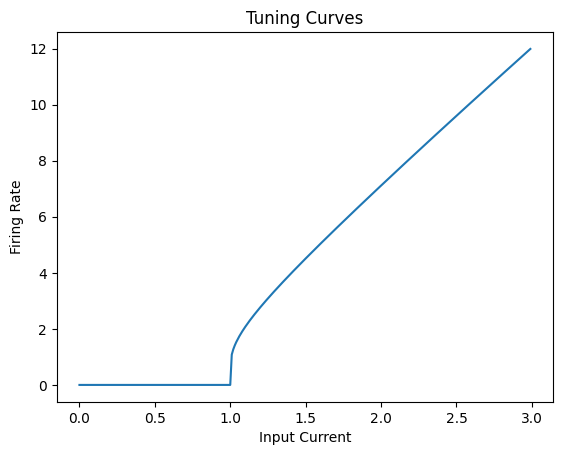
If we look at the tuning curve of a neuron (how it responds to different input values), we get:
Show code cell source
import matplotlib.pyplot as plt
import numpy as np
np.random.seed(0)
def computeTuningCurve(lif, inputs, time_limit = 1, t_step = 0.001):
tuningCurve = []
for inp in inputs:
spike_count = 0
lif.reset()
for _ in np.arange(0, time_limit, t_step):
output = lif.step(inp, t_step)
if output > 0:
spike_count += 1
rate = spike_count / time_limit
tuningCurve.append(rate)
lif.reset()
return tuningCurve
def analyticalRate(neuron, I):
if I <= neuron.v_th: return 0
else: return 1 / (neuron.tau_ref - neuron.tau_rc * np.log(1 - neuron.v_th/I))
neuron = FirstOrderLIF()
inputs = np.arange(0, 3, 0.01)
# tuningCurve = computeTuningCurve(neuron, inputs)
tuningCurve = [analyticalRate(neuron, i) for i in inputs]
plt.figure()
plt.plot(inputs, tuningCurve)
plt.xlabel('Input Current')
plt.ylabel('Firing Rate')
plt.title('Tuning Curves')
plt.show()
psp = FirstOrderSynapse(tau_s = 0.15)
def computeDecoder(neuron, range_low=-1, range_high=1, interval=0.1):
numerator = 0
denominator = 0
for i in np.arange(range_low, range_high, interval):
r = analyticalRate(neuron, i)
numerator += r*i
denominator += r*r
return numerator / denominator
# plot signal vs time for 12 seconds
t_step = 0.01
t = np.arange(0, 12, t_step)
decoder = computeDecoder(neuron, -2, 2)
print(f"The decoder is {decoder}")
signal_out = []
decoded_output = []
for i in t:
sig = signal(i)
neuron.step(sig, t_step)
psp.step(neuron.output, t_step)
signal_out.append(sig)
decoded_output.append(decoder * psp.output)
plt.figure()
plt.plot(t, decoded_output)
plt.plot(t, signal_out, linestyle="--")
plt.xlabel('Time (s)')
plt.ylabel('Output')
plt.title('Decoded Output')
plt.legend(['Decoded Output', 'Input Signal'])
plt.show()
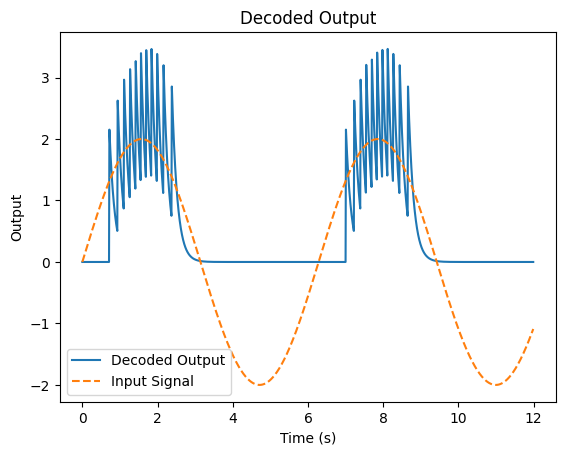
The decoder is 0.3229210069532396
So far, we have focused on single neurons but in order to convey information effectively, we need populations of neurons—not just one. In other words, we are going to move from using one neuron to using many neurons to better represent (and later on, process) our signals. If we work with a population of neurons, each neuron should react differently to the same input (meaning they should have different tuning curves). If two neurons had the exact same tuning curve, they aren’t conveying any more information than one neuron. This should convey more information than just one neuron, which we can verify by decoding the signals of multiple neurons.
Now that we’re working with multiple neurons, we will also start using the subscript \(_{i}\) to represent neuron \(i\). We’ll also add three additional terms to scale the input of every neuron:
\(\alpha_i\): the gain—a multiplicative factor that scales the input
\(e_i\): the encoder—a scale that decides what “direction” our neuron responds to (in our case, it will be either \(1\) or \(-1\) to specify if it fires more for positive or negative inputs?)
\(b_i\): the bias—an bias that we add to the input
This means that for input \(x\), the effective input for neuron \(i\) will be \(e_{i}\alpha_{i}x + b_i\).
Note
If \(e_{i}\) is negative, our neuron will fire more frequently for negative inputs. Although this does not necessarily make sense in the context of a single isolated neuron, (1) biological realism is not our primary goal and (2) it might correspond to a situation where there is some constant background current and an inhibitory background current.
It might also seem strange that we separate the gain \(\alpha_{i}\) from the encoder \(e_{i}\), with \(e\) representing the “direction” that our neuron responds to and \(\alpha\) representing the scale [Ste12]. It will make more sense to do this when we have multiple inputs and there’s a notion of “direction” (in other words, how does our neuron react to each input).
Since it can be a little easier to specify the “intercept” (\(x_{int}\)—the input where our neuron starts firing) and the maximum firing rate (\(r_{max}\)—how fast our neuron can fire for the inputs we expect), we can write a function (getGainBias) to compute the values of \(\alpha_i\) and \(b_i\) from \(r_{max}\), \(x_{int}\), and information about the neuron—\(\tau_{rc}\), \(\tau_{ref}\), and \(v_{th}\):
def getGainBias(maxRate, intercept, tau_rc, tau_ref, v_th):
gain = v_th * (1 - 1 / (1 - np.exp((tau_ref - 1/maxRate) / tau_rc))) / (intercept - 1)
bias = v_th - gain * intercept
return gain, bias
Where does this code/math for computing the gain and bias come from?
Computing Scales and Biases (Optional)
We could randomly select values of \(\alpha_i\) and \(b_i\) but let’s instead compute them from:
A desired maximum firing rate in the context of an interval, which we will call \(r_{max}\)
A desired intercept, where our neuron starts firing, which we will call \(x_{int}\)
We can analytically determine \(r(I)\) to estimate a firing rate (5):
We can turn this equation around to ask what input current do we need to achieve a given firing rate. In other words, we can leave \(r(I)\) fixed (at \(r\)) and solve for \(I\). If we do this on the above equation, we get:
Let’s call this \(z(r)\) for short:
Recall that our effective input given input \(x\) is \(I\):
Let’s suppose that we only care about the possible inputs up to \(1\). This means that our maximum rate, \(r_{max}\) should be achieved at input \(x=1\):
or re-arranging:
But we know that at our intercept \(x_{int}\), we should not fire, meaning that it is right at the threshold of our firing voltage, \(v_{th}\): \(I(x_{int}) = v_{th}\). So we have:
or re-arranging:
Putting both equations for \(b\) together, we get:
…re-arranging:
and
Let’s then write code for representing a collection of that creates a collection of neurons with different tuning curves
class FirstOrderLIFCollection: # Collection of First Order Leaky Integrate and Fire Neurons
def __init__(self, num_neurons, tau_rc=0.02, tau_ref=0.002, v_th=1, max_rate_range = (200, 400), intercept_range = (-1, 1), encoder_options = (-1, 1)):
self.neurons = [] # List of neurons
self.gains = [] # List of gains (numbers)
self.biases = [] # List of biases (numbers)
self.encoders = [] # List of encoders (numbers)
for _ in range(num_neurons):
neuron = FirstOrderLIF(tau_rc=tau_rc, tau_ref=tau_ref, v_th=v_th)
max_rate = np.random.uniform(max_rate_range[0], max_rate_range[1]) # Maximum firing rate
intercept = np.random.uniform(intercept_range[0], intercept_range[1]) # Intercept (where the neuron starts firing)
encoder = np.random.choice(encoder_options) # Encoder (direction of the neuron's tuning curve)
gain, bias = getGainBias(max_rate, intercept, tau_rc, tau_ref, v_th)
self.neurons.append(neuron)
self.gains.append(gain)
self.biases.append(bias)
self.encoders.append(encoder)
def step(self, I, t_step):
outputs = []
for neuron, gain, bias, encoder in zip(self.neurons, self.gains, self.biases, self.encoders): # Loop over neurons, gains, biases, and encoders
output = neuron.step(I * gain * encoder + bias, t_step) # Step the neuron with the input scaled by the gain and encoder, plus the bias
outputs.append(output) # Append the output to the list of outputs
return outputs
def reset(self): # Reset all neurons to their initial state
for neuron in self.neurons:
neuron.reset()
Let’s then create a collection of ten neurons with different tuning curves.
NUM_NEURONS = 15
lifs = FirstOrderLIFCollection(NUM_NEURONS, max_rate_range=(25, 100))
def computeTuningCurves(lifs, inputs, time_limit = 1, t_step = 0.001):
tuning_curves = [ [0] * len(inputs) for _ in range(len(lifs.neurons)) ]
num_neurons = len(lifs.neurons)
for inp_idx, inp in enumerate(inputs):
spike_counts = [0] * num_neurons
lifs.reset()
for _ in np.arange(0, time_limit, t_step):
outputs = lifs.step(inp, t_step)
for i in range(num_neurons):
if outputs[i] > 0:
spike_counts[i] += 1
for i in range(num_neurons):
tuning_curves[i][inp_idx] = spike_counts[i] / time_limit
lifs.reset()
return tuning_curves
inputs = np.arange(-1, 1, 0.01)
plt.figure()
# for curve in computeTuningCurves(lifs, inputs):
# plt.plot(inputs, curve)
for neuron, gain, bias, encoder in zip(lifs.neurons, lifs.gains, lifs.biases, lifs.encoders):
tuning_curve = [analyticalRate(neuron, i * gain * encoder + bias) for i in inputs]
plt.plot(inputs, tuning_curve)
plt.xlabel('Input Current')
plt.ylabel('Firing Rate')
plt.title('Tuning Curves')
plt.show()
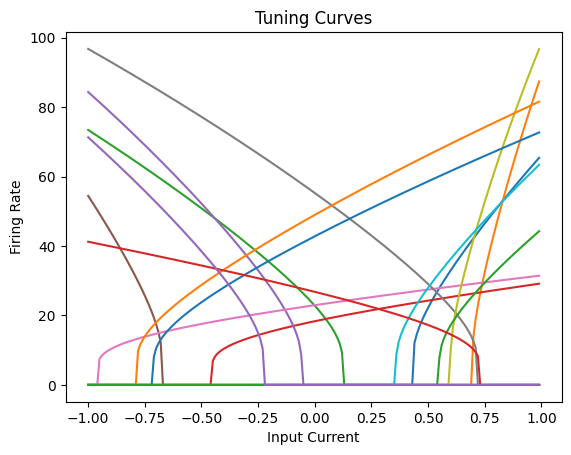
As we can see, all the neurons have different tuning curves as a result of their random gains, biases, and encoders.
Let’s then also define a FirstOrderSynapseCollection class that will represent a collection of synapses for these neurons.
class FirstOrderSynapseCollection:
def __init__(self, num_neurons, tau_s=0.05):
self.synapses = [FirstOrderSynapse(tau_s) for _ in range(num_neurons)]
def step(self, inputs, t_step):
outputs = []
for i, synapse in enumerate(self.synapses):
inp = inputs[i]
output = synapse.step(inp, t_step)
outputs.append(output)
return outputs
def reset(self):
for synapse in self.synapses:
synapse.reset()
Then, let’s create a collection of synapses and see what happens with a signal.
def signal(t):
return math.sin(t)
synapses = FirstOrderSynapseCollection(NUM_NEURONS, tau_s=0.1)
def step(lifs, synapses, input_value, t_step):
lif_outputs = lifs.step(input_value, t_step)
synapse_outputs = synapses.step(lif_outputs, t_step)
return synapse_outputs
T_step = 0.001
T = np.arange(0, 12, T_step)
signals = []
outputs = []
for t in T:
input_value = signal(t)
out = step(lifs, synapses, input_value, T_step)
signals.append(input_value)
outputs.append(out)
plt.figure()
plt.plot(T, signals, color="C1")
plt.xlabel('Time (s)')
plt.ylabel('Input Signal')
plt.show()
plt.figure()
plt.plot(T, outputs)
plt.xlabel('Time (s)')
plt.ylabel('Outputs')
plt.show()
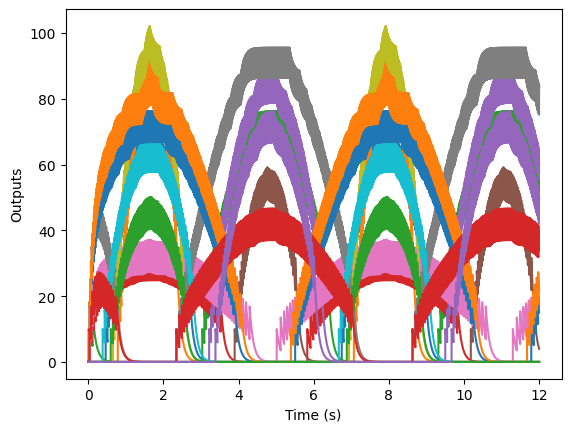
Now, if we want to try to re-construct the original signal from these neurons’ outputs, we need to compute the decoders for multiple neurons. The procedure is similar to what we did with one neuron, except that now we are minimizing the error across several neurons. So now, if we have \(n\) neurons, our decoders \(\vec{\phi}\) will be a vector rather than a single number. We use \(\phi_i\) to refer to the decoder for neuron \(i\).
What’s the math behind computing \(\vec{\phi}\)?
Computing Population Decoders (Optional)
When we want to compute the decoders for multiple neurons, the procedure is similar as it was for one neuron. However, now we are minimizing the error across several neurons. So now, if we have \(n\) neurons, our decoders \(\vec{\phi}\) will be a vector rather than a single number. We use \(\phi_i\) to refer to the decoder for neuron \(i\).
We could minimize this procedurally (for example, through gradient descent) or analytically. For large numbers of neurons, it’s probably best to do it procedurally but let’s instead do it analytically. We want to find the optimal values for each \(\phi_i\) by looking for the point where \(\frac{\partial{}E(j)}{\partial{}\phi_i} = 0\).
If we set this equal to zero, we have:
re-arranging
If we define matrices \(\Upsilon{}\) and \(\Gamma{}\) as follows:
and
then we get
or
We can estimate the integrals for \(\Gamma\) and \(\Upsilon\) through summation.
Two more notes:
Some sources recommend adding random noise to be more sure that \(\Gamma\) is invertible. They recommend adding \(\sigma{}\), the standard deviation of the noise to the diagonal so we will use \(\Gamma + I\) where \(I\) is the identity matrix.
Our computation of \(\vec{\phi}\) is slightly inaccurate, especially for large numbers of neurons because of some implementation details (the library we use for matrix inversion is not precise…the optional section below discusses why we use matrix inversion).
import numpy as np
def getDecoders(lifs, minJ=-1, maxJ=1, stepSize=0.01):
inputs = np.arange(minJ, maxJ, stepSize)
# tuningCurves = computeTuningCurves(lifs, inputs)
tuningCurves = []
for neuron, gain, bias, encoder in zip(lifs.neurons, lifs.gains, lifs.biases, lifs.encoders):
tuningCurves.append([analyticalRate(neuron, i * gain * encoder + bias) for i in inputs])
A = np.array(tuningCurves)
value = np.expand_dims(inputs, axis=1)
Gamma = A @ A.T + np.identity(len(lifs.neurons))
GammaInv = np.linalg.inv(Gamma)
Upsilon = A @ value
Phi = GammaInv @ Upsilon
return Phi
Phi = getDecoders(lifs)
print(f"Phi is: {Phi.flatten()}")
Phi is: [ 1.25173521e-05 1.82993317e-03 -5.10319914e-03 1.50262571e-03
-1.00303516e-03 2.88883235e-04 5.12202562e-03 -9.81927479e-03
-5.44357947e-04 4.34728325e-03 1.89612567e-03 2.78694925e-03
2.72645357e-04 1.10751640e-02 -8.00340690e-04]
We can use our weights \(\vec{\phi}\) to try to re-construct the original signal by taking the outputs (outputs) and doing matrix multiplication with \(\vec{\phi}\):
Show code cell source
plt.figure()
plt.plot(T, outputs @ Phi) # Multiply the outputs by the decoders with matrix multiplication
plt.plot(T, signals, linestyle='--')
plt.xlabel('Time (s)')
plt.ylabel('Signal')
plt.title('Signal vs Time')
plt.legend(['Decoded Signal', 'Input Signal'])
plt.show()
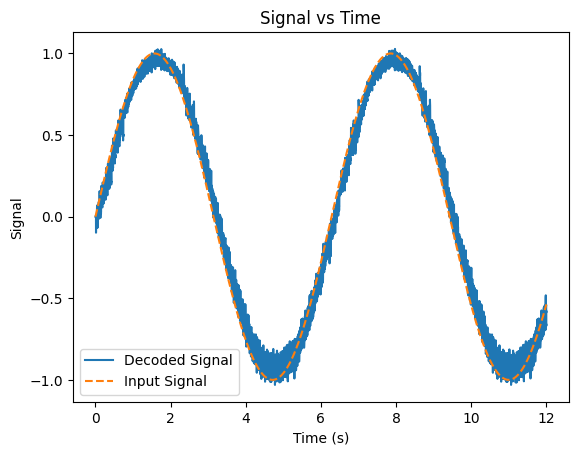
As we can see, even with a small number of neurons, we can re-construct our original input signal fairly well. If we added more neurons, our representation would improve.
Also, our time constant \(\tau_s\) has an interesting effect on our output. As we decrease its value, our representation gets a little “rougher”, since our PSP decays faster our representation ends up being jerkier. If we increase \(\tau_s\), we end up with a smoother representation but it becomes increasingly time-delayed and is slower to “react” to the input. The example below shows the output for a large value of \(\tau_s\) (prior value: 0.1, new value: 1.0):
Show code cell source
lifs = FirstOrderLIFCollection(NUM_NEURONS, max_rate_range=(25, 100))
synapses = FirstOrderSynapseCollection(NUM_NEURONS, tau_s=1)
signals = []
outputs = []
for t in T:
input_value = signal(t)
out = step(lifs, synapses, input_value, T_step)
signals.append(input_value)
outputs.append(out)
Phi = getDecoders(lifs)
plt.figure()
plt.plot(T, outputs @ Phi)
plt.plot(T, signals, linestyle='--')
plt.xlabel('Time (s)')
plt.ylabel('Signal')
plt.title('Signal vs Time')
plt.legend(['Decoded Signal', 'Input Signal'])
plt.show()
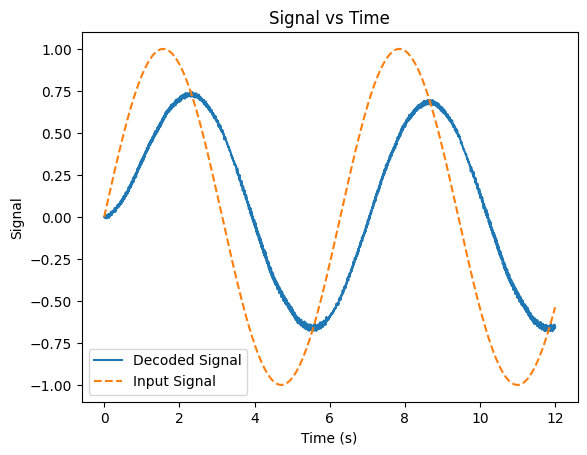
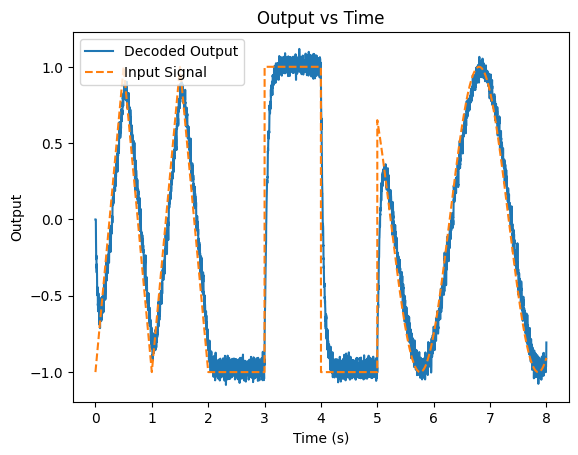
Let’s see what this looks like with a stranger signal (and up the number of neurons a bit to 50):
Show code cell source
lifs = FirstOrderLIFCollection(50, max_rate_range=(25, 100))
synapses = FirstOrderSynapseCollection(50)
def signal2(t): # Create a (strange) signal
if t < 2:
return 1 - 4 * abs((t%1) - 0.5) # Triangle wave for first two seconds
elif t < 5:
return -1 if (t % 2) < 1 else 1 # Square wave for the next three seconds
else:
return math.sin(t*3) # Sine wave for the remainder
T_step = 0.001
T = np.arange(0, 8, T_step)
signals = []
outputs = []
for t in T:
sig = signal2(t)
out = step(lifs, synapses, sig, T_step)
signals.append(sig)
outputs.append(out)
Phi = getDecoders(lifs)
decoded_output = outputs @ Phi
plt.figure()
plt.plot(T, decoded_output)
plt.plot(T, signals, linestyle="--")
plt.xlabel('Time (s)')
plt.ylabel('Output')
plt.title('Output vs Time')
plt.legend(['Decoded Output', 'Input Signal'])
plt.show()
Summary#
We can compute decoders to check that our neurons are able to re-construct an input signal
Decoders are generally used for debugging (for the humans creating the SNNs)
Our decoders are scalar values (which we called \(\phi{}\)) by which we multiply the post-synaptic potential to get an estimate of the input signal
We can create groups of neurons with different gains (\(\alpha{}\)), biases (\(b\)), and encoders (\(e\)) to better represent the input signal
Given input \(x\), the neuron’s effective input is: \(e\alpha{}x + b\)
We can then compute a decoder vector, \(\vec{\phi}\), to decode the original signal
Resources#
The code for computing scales and biases is adapted from Nengo’s implementation
Chris Eliasmith’s Neural Engineering book contains the derivation used for computing decoders
Nengo’s NEF Algorithm code is a good example of how to build and understand encoders
References#
Josh Achiam, Steven Adler, Sandhini Agarwal, Lama Ahmad, Ilge Akkaya, Florencia Leoni Aleman, Diogo Almeida, Janko Altenschmidt, Sam Altman, Shyamal Anadkat, and others. Gpt-4 technical report. arXiv preprint arXiv:2303.08774, 2023.
Peter U Diehl and Matthew Cook. Unsupervised learning of digit recognition using spike-timing-dependent plasticity. Frontiers in computational neuroscience, 9:99, 2015.
John McCarthy, Marvin L Minsky, Nathaniel Rochester, and Claude E Shannon. A proposal for the dartmouth summer research project on artificial intelligence, august 31, 1955. AI magazine, 27(4):12–12, 2006.
Terrence C Stewart. A technical overview of the neural engineering framework. University of Waterloo, 2012.