Weights and Connections#
When we have multiple groups of neurons, we then would want to connect them. This is done by assigning weights to the connections between the neurons, with the weights being the strength of the connection. For example, a weight of 0 would mean no connection, while a weight of 1 would mean a strong connection.
We can learn connection weights in a variety of ways, as we will go into later.
For now, we will initialize them randomly using np.random.randn(). This will give us a random number from a normal distribution with a mean of 0 and a standard deviation of 1.:
Show code cell source
import numpy as np
np.random.seed(1) # Set random seed for reproducibility
class LIFCollection:
def __init__(self, n=1, dim=1, tau_rc=0.02, tau_ref=0.002, v_th=1,
max_rates=[200, 400], intercept_range=[-1, 1], t_step=0.001, v_init = 0):
self.n = n
# Set neuron parameters
self.dim = dim # Dimensionality of the input
self.tau_rc = tau_rc # Membrane time constant
self.tau_ref = tau_ref # Refractory period
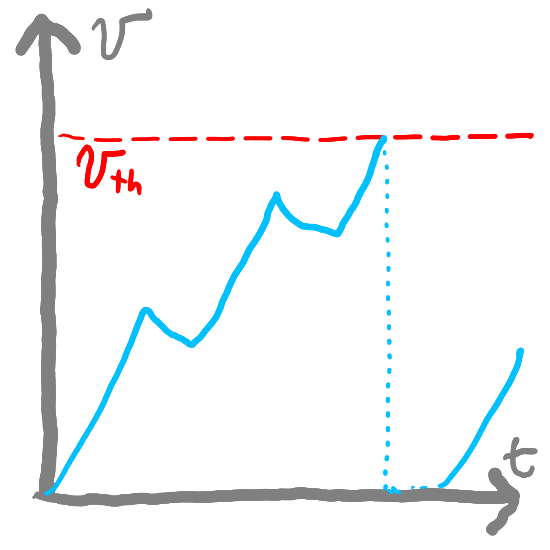
self.v_th = np.ones(n) * v_th # Threshold voltage for spiking
self.t_step = t_step # Time step for simulation
# Initialize state variables
self.voltage = np.ones(n) * v_init # Initial voltage of neurons
self.refractory_time = np.zeros(n) # Time remaining in refractory period
self.output = np.zeros(n) # Output spikes
# Generate random max rates and intercepts within the given range
max_rates_tensor = np.random.uniform(max_rates[0], max_rates[1], n)
intercepts_tensor = np.random.uniform(intercept_range[0], intercept_range[1], n)
# Calculate gain and bias for each neuron
self.gain = self.v_th * (1 - 1 / (1 - np.exp((self.tau_ref - 1/max_rates_tensor) / self.tau_rc))) / (intercepts_tensor - 1)
self.bias = np.expand_dims(self.v_th - self.gain * intercepts_tensor, axis=1)
# Initialize random encoders
self.encoders = np.random.randn(n, self.dim)
self.encoders /= np.linalg.norm(self.encoders, axis=1)[:, np.newaxis]
def reset(self):
# Reset the state variables to initial conditions
self.voltage = np.zeros(self.n)
self.refractory_time = np.zeros(self.n)
self.output = np.zeros(self.n)
def step(self, inputs):
dt = self.t_step # Time step
# Update refractory time
self.refractory_time -= dt
delta_t = np.clip(dt - self.refractory_time, 0, dt) # ensure between 0 and dt
# Calculate input current
I = np.sum(self.bias + inputs * self.encoders * self.gain[:, np.newaxis], axis=1)
# Update membrane potential
self.voltage = I + (self.voltage - I) * np.exp(-delta_t / self.tau_rc)
# Determine which neurons spike
spike_mask = self.voltage > self.v_th
self.output[:] = spike_mask / dt # Record spikes in output
# Calculate the time of the spike
t_spike = self.tau_rc * np.log((self.voltage[spike_mask] - I[spike_mask]) / (self.v_th[spike_mask] - I[spike_mask])) + dt
# Reset voltage of spiking neurons
self.voltage[spike_mask] = 0
# Set refractory time for spiking neurons
self.refractory_time[spike_mask] = self.tau_ref + t_spike
return self.output # Return the output spikes
class SynapseCollection:
def __init__(self, n=1, tau_s=0.05, t_step=0.001):
self.n = n
self.a = np.exp(-t_step / tau_s) # Decay factor for synaptic current
self.b = 1 - self.a # Scale factor for input current
self.voltage = np.zeros(n) # Initial voltage of neurons
def step(self, inputs):
self.voltage = self.a * self.voltage + self.b * inputs
return self.voltage
t_step = 0.001
neurons_a = LIFCollection(n=50, tau_rc=0.02, tau_ref=0.002, t_step=t_step)
synapses_a = SynapseCollection(n=neurons_a.n, tau_s=0.1, t_step=t_step)
neurons_b = LIFCollection(n=40, tau_rc=0.02, tau_ref=0.002, t_step=t_step)
synapses_b = SynapseCollection(n=neurons_b.n, tau_s=0.1, t_step=t_step)
weights = np.random.randn(neurons_a.n, neurons_b.n)
outp = []
def step(inp):
a = neurons_a.step(inp)
b = synapses_a.step(a)
bw = b @ weights
c = neurons_b.step(bw)
d = synapses_b.step(c)
return (a, b, bw, c, d)
T = 10
times = np.arange(0, T, t_step)
def inp(t):
return np.sin(t)
for t in times:
outp.append(step(inp(t)))